Exporting Crosslinks to AlphaLink2
from pyXLMS import __version__
print(f"Installed pyXLMS version: {__version__}") Installed pyXLMS version: 1.6.0from pyXLMS import parser
from pyXLMS import exporterAll exporting functionality is available via the exporter submodule. We also import the parser submodule to read crosslinks.
parser_result = parser.read(
"../../data/ms_annika/XLpeplib_Beveridge_QEx-HFX_DSS_R1_Crosslinks.xlsx",
engine="MS Annika",
crosslinker="DSS",
)
xls = parser_result["crosslinks"] Reading MS Annika crosslinks...: 100%|██████████████████████████████████████████████████████████████████████████| 300/300 [00:00<00:00, 19063.00it/s]We read crosslinks using the generic parser from a single MS Annika .xlsx file. For easier access we also assign our crosslinks to the variable xls.
from pyXLMS.transform import validate, targets_only
xls = targets_only(validate(xls)) Iterating over scores for FDR calculation...: 25%|██████████████████ | 74/300 [00:00<?, ?it/s]from pyXLMS.transform import filter_proteins
cas9 = filter_proteins(xls, proteins=["Cas9"])["Both"]You also should filter your crosslinks to only contain residue pairs of your protein(s) of interest, for example with the filter_proteins() [docs ] function!
result = exporter.to_alphalink2(
cas9,
fasta="../../data/_fasta/Cas9_plus10.fasta",
annotated_fdr=0.01,
filename_prefix="Cas9",
) Exporting crosslinks to AlphaLink2...: 100%|███████████████████████████████████████████████████████████████████████████████| 223/223 [00:00<?, ?it/s]The function exporter.to_alphalink2() exports a list of crosslinks to AlphaLink2 format for protein (complex) structure prediction with crosslink restraints. The tool AlphaLink2 is available via the link github.com/Rappsilber-Laboratory/AlphaLink2 . The function to_alphalink2() takes in a list of crosslinks as input and additionally needs a FASTA file as input, containing the sequences of the protein/chains of interest that should be modelled by AlphaLink2. Furthermore, the AlphaLink2 format requires an FDR column for your crosslinks. This is controlled via the annotated_fdr parameter which accepts a single float as input or a list of float values. In our case we specified annotated_fdr=0.01 which means that all crosslinks are labelled with a constant 1% false-discovery-rate (FDR). We could also pass a list of float values of equal length as our crosslinks to set individual FDR values for all crosslinks. You can read more about the to_alphalink2() function and all its parameters here: docs.
If transform.annotate_fdr() [docs ] has been run previously, the to_alphalink2() function will automatically use the pyXLMS annotated FDR for the AlphaLink2 FDR column and only use the parameter annotated_fdr as a backup. To override this behaviour pass try_use_annotated_fdr=False to the to_alphalink2() function which will explicitly use the annotated_fdr parameter value(s)!
for key, value in result.items():
print(f"{key}: {type(value)}") AlphaLink2 crosslinks: <class 'str'>
AlphaLink2 FASTA: <class 'str'>
AlphaLink2 DataFrame: <class 'pandas.core.frame.DataFrame'>
Exported files: <class 'list'>The exporter function returns a dictionary
- with key
"AlphaLink2 crosslinks"containing the formatted crosslink input for AlphaLink2, - with key
"AlphaLink2 FASTA"containing the FASTA file content for AlphaLink2, - with key
"AlphaLink2 DataFrame"containing the exported crosslinks as a pandas DataFrame , - and with key
"Exported files"containing a list of filenames of all files that were written to disk.
print("\n".join(result["AlphaLink2 crosslinks"].split("\n")[:5])) 779 A 779 A 0.01
866 A 866 A 0.01
677 A 677 A 0.01
677 A 48 A 0.01
34 A 34 A 0.01Via "AlphaLink2 crosslinks" we can access the formatted crosslink input for AlphaLink2, if filename_prefix was not None this has also been written to a file with name {filename_prefix}_AlphaLink2.txt.
print(result["AlphaLink2 FASTA"]) >A|Cas9
GAASMDKKYSIGLAIGTNSVGWAVITDEYKVPSKKFKVLGNTDRHSIKKNLIGALLFDSGETAEATRLKRTARRRYTRRKNRICYLQEIFSNEMAKVDDSFFHRLEESFLVEEDKKHERHPIFGNIVDEVAYHEKYPTIYHLRKKLVDSTDKADLRLIYLALAHMIKFRGHFLIEGDLNPDNSDVDKLFIQLVQTYNQLFEENPINASGVDAKAILSARLSKSRRLENLIAQLPGEKKNGLFGNLIALSLGLTPNFKSNFDLAEDAKLQLSKDTYDDDLDNLLAQIGDQYADLFLAAKNLSDAILLSDILRVNTEITKAPLSASMIKRYDEHHQDLTLLKALVRQQLPEKYKEIFFDQSKNGYAGYIDGGASQEEFYKFIKPILEKMDGTEELLVKLNREDLLRKQRTFDNGSIPHQIHLGELHAILRRQEDFYPFLKDNREKIEKILTFRIPYYVGPLARGNSRFAWMTRKSEETITPWNFEEVVDKGASAQSFIERMTNFDKNLPNEKVLPKHSLLYEYFTVYNELTKVKYVTEGMRKPAFLSGEQKKAIVDLLFKTNRKVTVKQLKEDYFKKIECFDSVEISGVEDRFNASLGTYHDLLKIIKDKDFLDNEENEDILEDIVLTLTLFEDREMIEERLKTYAHLFDDKVMKQLKRRRYTGWGRLSRKLINGIRDKQSGKTILDFLKSDGFANRNFMQLIHDDSLTFKEDIQKAQVSGQGDSLHEHIANLAGSPAIKKGILQTVKVVDELVKVMGRHKPENIVIEMARENQTTQKGQKNSRERMKRIEEGIKELGSQILKEHPVENTQLQNEKLYLYYLQNGRDMYVDQELDINRLSDYDVDAIVPQSFLKDDSIDNKVLTRSDKNRGKSDNVPSEEVVKKMKNYWRQLLNAKLITQRKFDNLTKAERGGLSELDKAGFIKRQLVETRQITKHVAQILDSRMNTKYDENDKLIREVKVITLKSKLVSDFRKDFQFYKVREINNYHHAHDAYLNAVVGTALIKKYPKLESEFVYGDYKVYDVRKMIAKSEQEIGKATAKYFFYSNIMNFFKTEITLANGEIRKRPLIETNGETGEIVWDKGRDFATVRKVLSMPQVNIVKKTEVQTGGFSKESILPKRNSDKLIARKKDWDPKKYGGFDSPTVAYSVLVVAKVEKGKSKKLKSVKELLGITIMERSSFEKNPIDFLEAKGYKEVKKDLIIKLPKYSLFELENGRKRMLASAGELQKGNELALPSKYVNFLYLASHYEKLKGSPEDNEQKQLFVEQHKHYLDEIIEQISEFSKRVILADANLDKVLSAYNKHRDKPIREQAENIIHLFTLTNLGAPAAFKYFDTTIDRKQYRSTKEVLDATLIHQSITGLYETRIDLSQLGGDVia "AlphaLink2 FASTA" we can access the FASTA file content for AlphaLink2, if filename_prefix was not None this has also been written to a file with name {filename_prefix}_AlphaLink2.fasta.
result["AlphaLink2 DataFrame"].head()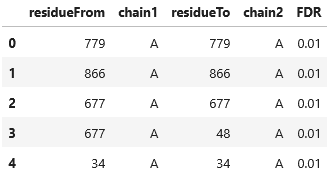
Via "AlphaLink2 DataFrame" we can access the crosslink input for AlphaLink2 as a pandas DataFrame .
result["Exported files"] ['Cas9_AlphaLink2.txt', 'Cas9_AlphaLink2.fasta']Via "Exported files" we can access the names of all files that were written to disk.
Using the AlphaLink2 Exporter with transform.annotate_fdr()
In order to retrieve more accurate false-discovery-rate (FDR) estimates for the FDR column in the AlphaLink2 crosslink table we can use the pyXLMS function transform.annotate_fdr() [docs , page] which will calculate FDR estimates for all crosslinks.
parser_result = parser.read(
"../../data/ms_annika/XLpeplib_Beveridge_QEx-HFX_DSS_R1_Crosslinks.xlsx",
engine="MS Annika",
crosslinker="DSS",
)
xls = parser_result["crosslinks"] Reading MS Annika crosslinks...: 100%|██████████████████████████████████████████████████████████████████████████| 300/300 [00:00<00:00, 18977.32it/s]We read crosslinks using the generic parser from a single MS Annika .xlsx file. For easier access we also assign our crosslinks to the variable xls.
from pyXLMS.transform import annotate_fdr
xls = annotate_fdr(xls) Annotating FDR for crosslinks...: 100%|████████████████████████████████████████████████████████████████████████████████████| 300/300 [00:00<?, ?it/s]from pyXLMS.transform import validate, targets_only
xls = targets_only(validate(xls)) Iterating over scores for FDR calculation...: 25%|██████████████████ | 74/300 [00:00<?, ?it/s]from pyXLMS.transform import filter_proteins
cas9 = filter_proteins(xls, proteins=["Cas9"])["Both"]You also should filter your crosslinks to only contain residue pairs of your protein(s) of interest, for example with the filter_proteins() [docs ] function!
result = exporter.to_alphalink2(
cas9,
fasta="../../data/_fasta/Cas9_plus10.fasta",
annotated_fdr=0.01,
try_use_annotated_fdr=True,
filename_prefix=None,
) Exporting crosslinks to AlphaLink2...: 100%|███████████████████████████████████████████████████████████████████████████████| 223/223 [00:00<?, ?it/s]The function exporter.to_alphalink2() exports a list of crosslinks to AlphaLink2 format for protein (complex) structure prediction with crosslink restraints. The tool AlphaLink2 is available via the link github.com/Rappsilber-Laboratory/AlphaLink2 . The function to_alphalink2() takes in a list of crosslinks as input and additionally needs a FASTA file as input, containing the sequences of the protein/chains of interest that should be modelled by AlphaLink2. Furthermore, the AlphaLink2 format requires an FDR column for your crosslinks. This is controlled via the annotated_fdr parameter which accepts a single float as input or a list of float values. In our case we specified annotated_fdr=0.01 which means that all crosslinks are labelled with a constant 1% false-discovery-rate (FDR). We could also pass a list of float values of equal length as our crosslinks to set individual FDR values for all crosslinks. However, because we previously annotated the FDR for our crosslinks with transform.annotate_fdr() the value(s) passed via parameter annotated_fdr are only used as a backup, e.g. if the FDR calculated by transform.annotate_fdr() is nan. This behaviour is controlled by the parameter try_use_annotated_fdr=True which is by default True. You can read more about the to_alphalink2() function and all its parameters here: docs.
Specifying filename_prefix=None will only return the calculated results but not write them to disk!
result["AlphaLink2 DataFrame"].head()
The FDR column now corresponds to the values that were calculated by transform.annotate_fdr(), even though we passed annotated_fdr=0.01.