Annotating Estimated False-Discovery-Rate
from pyXLMS import __version__
print(f"Installed pyXLMS version: {__version__}") Installed pyXLMS version: 1.6.0from pyXLMS import parser
from pyXLMS import transformAll data transformation functionality - including annotate_fdr() - is available via the transform submodule. We also import the parser submodule here for reading result files.
parser_result = parser.read(
"../../data/ms_annika/XLpeplib_Beveridge_QEx-HFX_DSS_R1.pdResult",
engine="MS Annika",
crosslinker="DSS",
) Reading MS Annika CSMs...: 100%|████████████████████████████████████████████████████████████████████████████████| 826/826 [00:00<00:00, 10258.12it/s]
Reading MS Annika crosslinks...: 100%|██████████████████████████████████████████████████████████████████████████| 300/300 [00:00<00:00, 20267.23it/s]We read crosslink-spectrum-matches and crosslinks using the generic parser from a single .pdResult file.
csms = parser_result["crosslink-spectrum-matches"]
xls = parser_result["crosslinks"]For easier access we assign our crosslink-spectrum-matches to the variable csms and our crosslinks to the variable xls.
Introduction
pyXLMS offers estimated false-discovery-rate (FDR) annotation for crosslink-spectrum-matches and crosslinks via the transform.annotate_fdr() function. Since this function shares a similar interface to transform.validate() [docs , page] we also recommend the reader to get familiar with that function!
Please note that FDR annotation is not validation! If you are looking for FDR-based validation please use transform.validate() [docs , page] instead! FDR annotation will associate an estimated FDR value to all crosslink-spectrum-matches or crosslinks and is significantly slower (O(n^2) runtime) than validation!
Before we run any kind of examples, let’s have a look at the function and its parameters:
def annotate_fdr(
data: List[Dict[str, Any]] | Dict[str, Any],
formula: Literal["D/T", "(TD+DD)/TT", "(TD-DD)/TT"] = "D/T",
score: Literal["higher_better", "lower_better"] = "higher_better",
separate_intra_inter: bool = False,
ignore_missing_labels: bool = False,
) -> List[Dict[str, Any]] | Dict[str, Any]:The function has 5 possible parameters:
- data: A list of crosslink-spectrum-matches or crosslinks to annotate, or a
parser_result. - formula: One of
"D/T","(TD+DD)/TT", and"(TD-DD)/TT"denoting which formula to use to estimate FDR.Ddenotes any decoy matches including decoy-decoy, decoy-target and target-decoy matches.DDdenotes decoy-decoy matches,TandTTdenote target-target matches, andTDdenotes target-decoy and decoy-target matches. - score: One of
"higher_better", or"lower_better"denoting if a higher score is considered better, or a lower score is considered better. - separate_intra_inter: If FDR should be estimated separately for intra and inter matches.
- ignore_missing_labels: If crosslinks and crosslink-spectrum-matches should be ignored if they don’t have target and decoy labels. By default and error is thrown if any unlabelled data is encountered.
Except for data all other parameters are optional with sensible defaults in place to stricly estimate FDR and annotate results. The data parameter supports lists of crosslink-spectrum-matches or crosslinks as input, or a complete parser_result. If a list of crosslink-spectrum-matches was provided, annotated_fdr() will return a list of FDR annotated crosslink-spectrum-matches. If a list of crosslinks was provided, annotate_fdr() will return a list of FDR annotated crosslinks. If a parser_result was provided, annotate_fdr() will return a parser_result where all elements are FDR annotated.
Please note that the by default selected FDR formula of ‘D/T’ actually denotes ‘(TD+DD)/TT’ where ‘TD’ includes both ‘TD’ and ‘DT’ matches since we consider any peptide pair with at least one decoy hit a decoy. Estimating FDR via ‘DD/TT’ is not valid for crosslinking mass spectrometry as it severly underestimated the actual error and is therefore not supported by pyXLMS!
Please note that FDR annotation in pyXLMS uses a generic target-decoy approach with a single score to estimate FDR. This might not be reflective of the FDR calculated by the used crosslink search engine! We also recommend reading the limitations of the specific crosslink search engine result parsers in the API docs .
FDR Annotation of Crosslink-Spectrum-Matches
_ = transform.summary(csms) Number of CSMs: 826.0
Number of unique CSMs: 826.0
Number of intra CSMs: 803.0
Number of inter CSMs: 23.0
Number of target-target CSMs: 786.0
Number of target-decoy CSMs: 39.0
Number of decoy-decoy CSMs: 1.0
Minimum CSM score: 1.1132827525593785
Maximum CSM score: 452.9861536355926Before annotation, let’s have a look at our crosslink-spectrum-matches using the transform.summary() function which you can read more about here: docs.
csms = transform.annotate_fdr(csms) Annotating FDR for crosslink-spectrum-matches...: 100%|█████████████████████████████████████████████████████████| 826/826 [00:00<00:00, 26426.76it/s]We can annotate our CSMs using the transform.annotate_fdr() function. For strict FDR annotation we can leave all optional parameters at their default values. You can read more about the annotate_fdr() function and all its parameters here: docs.
_ = transform.summary(csms) Number of CSMs: 826.0
Number of unique CSMs: 826.0
Number of intra CSMs: 803.0
Number of inter CSMs: 23.0
Number of target-target CSMs: 786.0
Number of target-decoy CSMs: 39.0
Number of decoy-decoy CSMs: 1.0
Minimum CSM score: 1.1132827525593785
Maximum CSM score: 452.9861536355926df = transform.to_dataframe(csms)
df["pyXLMS annotated FDR"] = [
csm["additional_information"]["pyXLMS_annotated_FDR"] for csm in csms
]
df.head()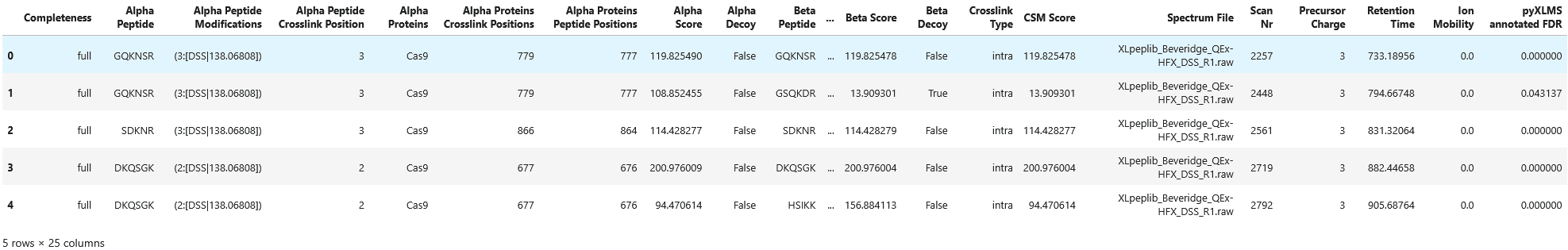
As a result we get the same list of crosslink-spectrum-matches that is now annotated with their FDR via strict FDR estimation.
csms = transform.annotate_fdr(csms, formula="(TD-DD)/TT", separate_intra_inter=True)
df = transform.to_dataframe(csms)
df["pyXLMS annotated FDR"] = [
csm["additional_information"]["pyXLMS_annotated_FDR"] for csm in csms
]
df.head() Annotating FDR for crosslink-spectrum-matches...: 100%|█████████████████████████████████████████████████████████| 803/803 [00:00<00:00, 26310.85it/s]
Annotating FDR for crosslink-spectrum-matches...: 100%|██████████████████████████████████████████████████████████████████████| 23/23 [00:00<?, ?it/s]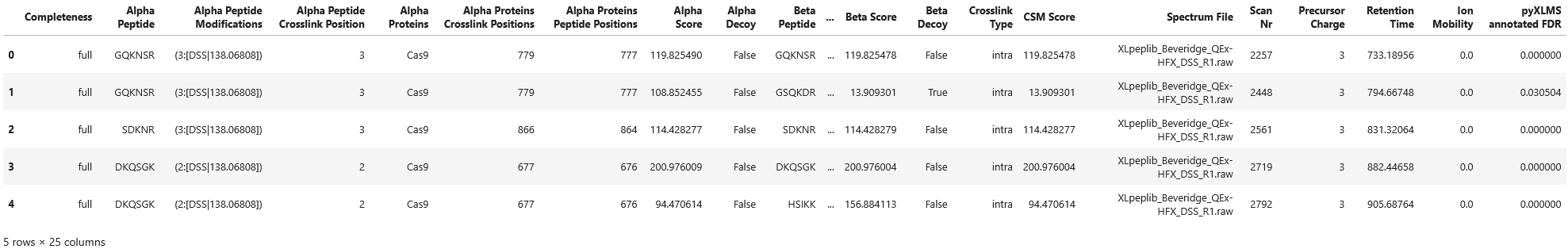
Of course we can also do more relaxed FDR estimation and annotation using a different formula formula="(TD-DD)/TT". We also separate FDR estimation by intra and inter matches separate_intra_inter=True.
FDR Annotation of Crosslinks
_ = transform.summary(xls) Number of crosslinks: 300.0
Number of unique crosslinks by peptide: 300.0
Number of unique crosslinks by protein: 298.0
Number of intra crosslinks: 279.0
Number of inter crosslinks: 21.0
Number of target-target crosslinks: 265.0
Number of target-decoy crosslinks: 0.0
Number of decoy-decoy crosslinks: 35.0
Minimum crosslink score: 1.1132827525593785
Maximum crosslink score: 452.9861536355926Before annotation, let’s have a look at our non-validated crosslinks using the transform.summary() function which you can read more about here: docs.
xls = transform.annotate_fdr(xls) Annotating FDR for crosslinks...: 100%|█████████████████████████████████████████████████████████████████████████| 300/300 [00:00<00:00, 75005.44it/s]Similarly to annotating CSMs, we can annotate our crosslinks using the transform.annotate_fdr() function. For strict FDR annotation we can leave all optional parameters at their default values. You can read more about the annotate_fdr() function and all its parameters here: docs.
_ = transform.summary(xls) Number of crosslinks: 300.0
Number of unique crosslinks by peptide: 300.0
Number of unique crosslinks by protein: 298.0
Number of intra crosslinks: 279.0
Number of inter crosslinks: 21.0
Number of target-target crosslinks: 265.0
Number of target-decoy crosslinks: 0.0
Number of decoy-decoy crosslinks: 35.0
Minimum crosslink score: 1.1132827525593785
Maximum crosslink score: 452.9861536355926df = transform.to_dataframe(xls)
df["pyXLMS annotated FDR"] = [
xl["additional_information"]["pyXLMS_annotated_FDR"] for xl in xls
]
df.head()
As a result we get the same list of crosslinks that is now annotated with their FDR via strict FDR estimation.
xls = transform.annotate_fdr(xls, separate_intra_inter=True)
df = transform.to_dataframe(xls)
df["pyXLMS annotated FDR"] = [
xl["additional_information"]["pyXLMS_annotated_FDR"] for xl in xls
]
df.head() Annotating FDR for crosslinks...: 100%|█████████████████████████████████████████████████████████████████████████| 279/279 [00:00<00:00, 79557.47it/s]
Annotating FDR for crosslinks...: 100%|██████████████████████████████████████████████████████████████████████████████████████| 21/21 [00:00<?, ?it/s]
Of course we can also do more relaxed FDR estimation and annotation, here using separate FDR estimation for intra and inter matches separate_intra_inter=True.
FDR Annotation of a parser_result
parser_result = transform.annotate_fdr(parser_result) Annotating FDR for crosslink-spectrum-matches...: 100%|█████████████████████████████████████████████████████████| 826/826 [00:00<00:00, 25660.27it/s]
Annotating FDR for crosslinks...: 100%|█████████████████████████████████████████████████████████████████████████| 300/300 [00:00<00:00, 74978.62it/s]We can even annotate our complete parser_result using the transform.annotate_fdr() function as well. For annotation using a strict FDR estimation we can also leave all optional parameters at their default values. You can read more about the annotate_fdr() function and all its parameters here: docs.